library(Canek)
# Functions
## Function to plot the pca coordinates
plotPCA <- function(pcaData = NULL, label = NULL, legPosition = "topleft"){
col <- as.integer(label)
plot(x = pcaData[,"PC1"], y = pcaData[,"PC2"],
col = as.integer(label), cex = 0.75, pch = 19,
xlab = "PC1", ylab = "PC2")
legend(legPosition, pch = 19,
legend = levels(label),
col = unique(as.integer(label)))
}Load the data
On this toy example we use the two simulated batches included in the
SimBatches data from Canek’s package.
SimBatches is a list containing:
-
batches: Simulated scRNA-seq datasets with genes (rows) and cells (columns). Simulations were performed using Splatter. -
cell_type: a factor containing the celltype labels of the batches
lsData <- list(B1 = SimBatches$batches[[1]], B2 = SimBatches$batches[[2]])
batch <- factor(c(rep("Batch-1", ncol(lsData[[1]])),
rep("Batch-2", ncol(lsData[[2]]))))
celltype <- SimBatches$cell_types
table(batch)
#> batch
#> Batch-1 Batch-2
#> 631 948
table(celltype)
#> celltype
#> Cell Type 1 Cell Type 2 Cell Type 3 Cell Type 4
#> 1451 53 38 37PCA before correction
We perform the Principal Component Analysis (PCA) of the joined datasets and scatter plot the first two PCs. The batch-effect causes cells to group by batch.
plotPCA(pcaData = pcaData, label = batch, legPosition = "bottomleft")
plotPCA(pcaData = pcaData, label = celltype, legPosition = "bottomleft")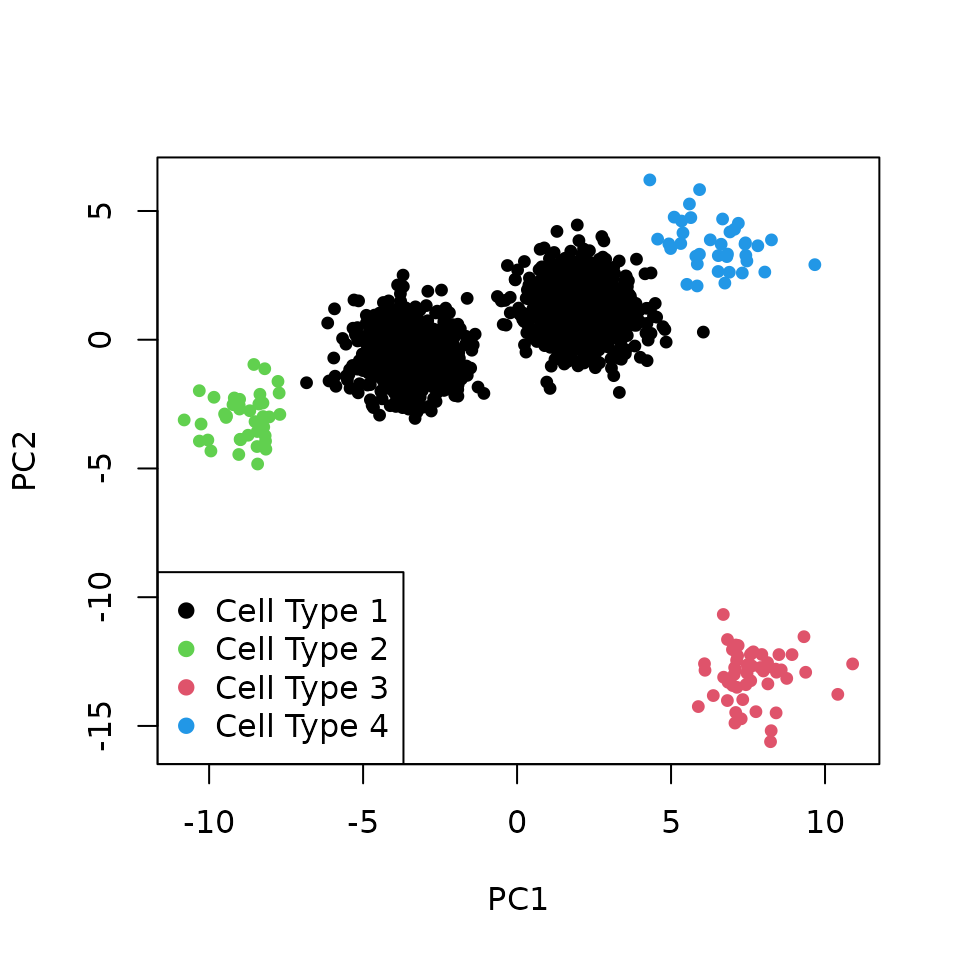
Run Canek
We correct the toy batches using the function RunCanek. This function accepts:
- List of matrices
- Seurat object
- List of Seurat objects
- SingleCellExperiment object
- List of SingleCellExperiment objects
On this example we use the list of matrices created before.
data <- RunCanek(lsData)PCA after correction
We perform PCA of the corrected datasets and plot the first two PCs. After correction, the cells group by their corresponding cell type.
plotPCA(pcaData = pcaData, label = batch, legPosition = "topleft")
plotPCA(pcaData = pcaData, label = celltype, legPosition = "topleft")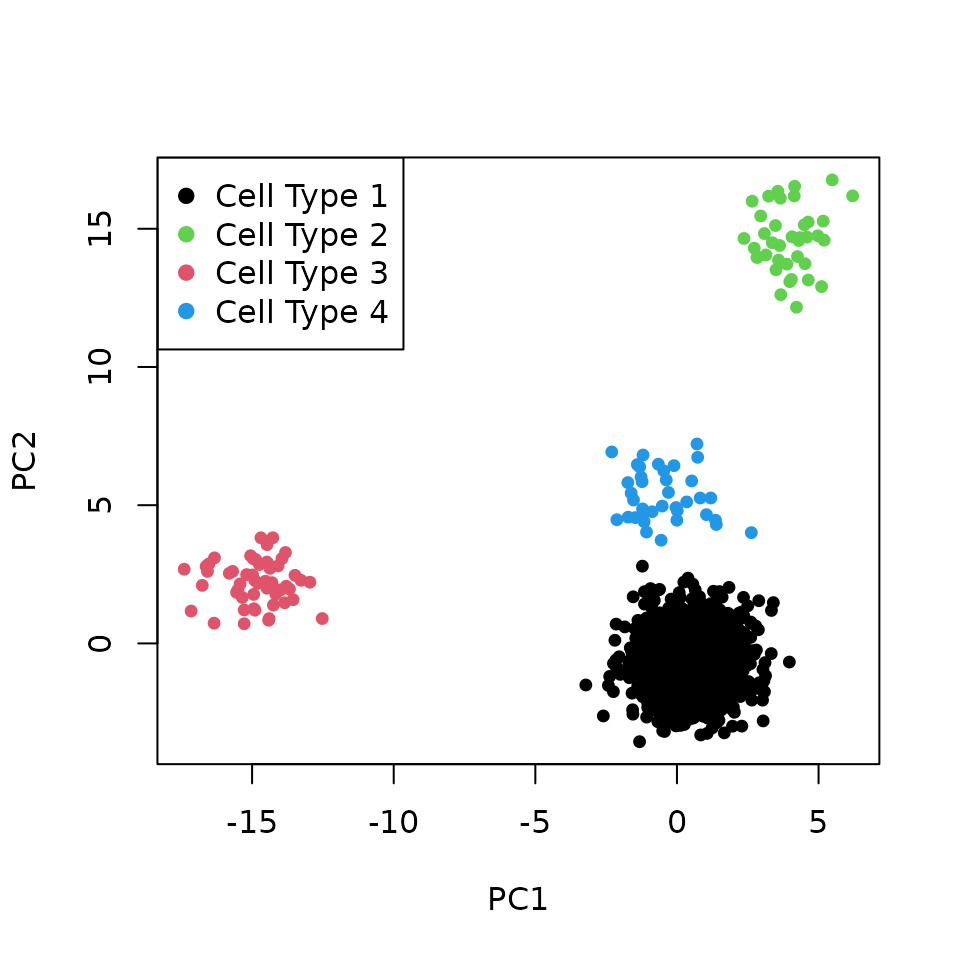
Session info
sessionInfo()
#> R version 4.3.2 (2023-10-31)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 22.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] Canek_0.2.4
#>
#> loaded via a namespace (and not attached):
#> [1] sass_0.4.7 robustbase_0.99-0 class_7.3-22
#> [4] stringi_1.7.12 lattice_0.21-9 numbers_0.8-5
#> [7] digest_0.6.33 magrittr_2.0.3 evaluate_0.23
#> [10] grid_4.3.2 fastmap_1.1.1 rprojroot_2.0.3
#> [13] jsonlite_1.8.7 Matrix_1.6-1.1 nnet_7.3-19
#> [16] mclust_6.0.0 kernlab_0.9-32 purrr_1.0.2
#> [19] codetools_0.2-19 modeltools_0.2-23 textshaping_0.3.7
#> [22] jquerylib_0.1.4 cli_3.6.1 rlang_1.1.1
#> [25] BiocNeighbors_1.18.0 cachem_1.0.8 yaml_2.3.7
#> [28] fpc_2.2-10 FNN_1.1.3.2 tools_4.3.2
#> [31] flexmix_2.3-19 parallel_4.3.2 BiocParallel_1.34.2
#> [34] memoise_2.0.1 BiocGenerics_0.46.0 vctrs_0.6.4
#> [37] R6_2.5.1 matrixStats_1.0.0 stats4_4.3.2
#> [40] lifecycle_1.0.3 stringr_1.5.0 S4Vectors_0.38.2
#> [43] fs_1.6.3 bluster_1.10.0 MASS_7.3-60
#> [46] irlba_2.3.5.1 ragg_1.2.6 cluster_2.1.4
#> [49] pkgconfig_2.0.3 desc_1.4.2 pkgdown_2.0.7
#> [52] bslib_0.5.1 glue_1.6.2 Rcpp_1.0.11
#> [55] systemfonts_1.0.5 highr_0.10 DEoptimR_1.1-3
#> [58] xfun_0.41 prabclus_2.3-3 knitr_1.45
#> [61] htmltools_0.5.6.1 igraph_1.5.1 rmarkdown_2.25
#> [64] compiler_4.3.2 diptest_0.76-0