library(Canek)
library(Seurat)
#> Attaching SeuratObject
#> 'SeuratObject' was built under R 4.3.0 but the current version is
#> 4.3.2; it is recomended that you reinstall 'SeuratObject' as the ABI
#> for R may have changed
#> Seurat v4 was just loaded with SeuratObject v5; disabling v5 assays and
#> validation routines, and ensuring assays work in strict v3/v4
#> compatibility modeCreate Seurat object
x <- lapply(names(SimBatches$batches), function(batch) {
CreateSeuratObject(SimBatches$batches[[batch]], project = batch)
})
x <- merge(x[[1]], x[[2]])
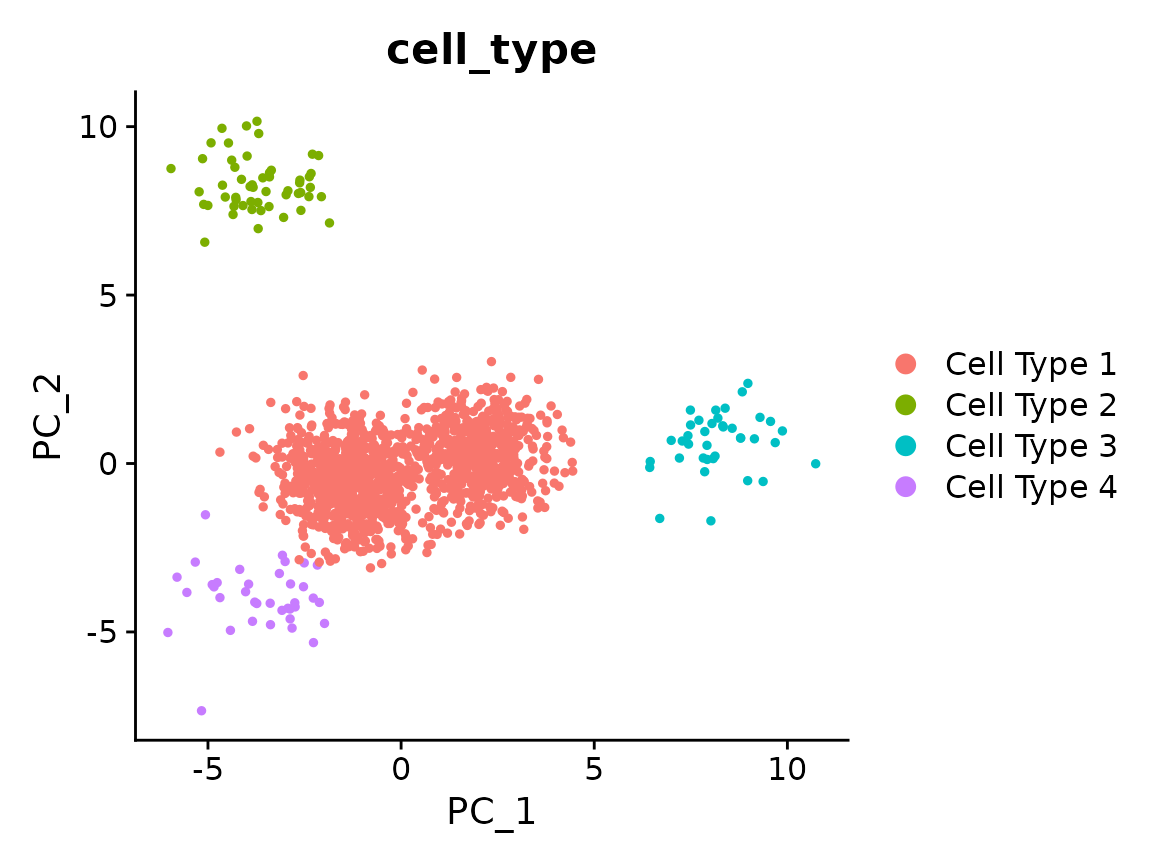
x[["cell_type"]] <- SimBatches$cell_types
x
#> An object of class Seurat
#> 500 features across 1579 samples within 1 assay
#> Active assay: RNA (500 features, 0 variable features)
#> 2 layers present: counts, data
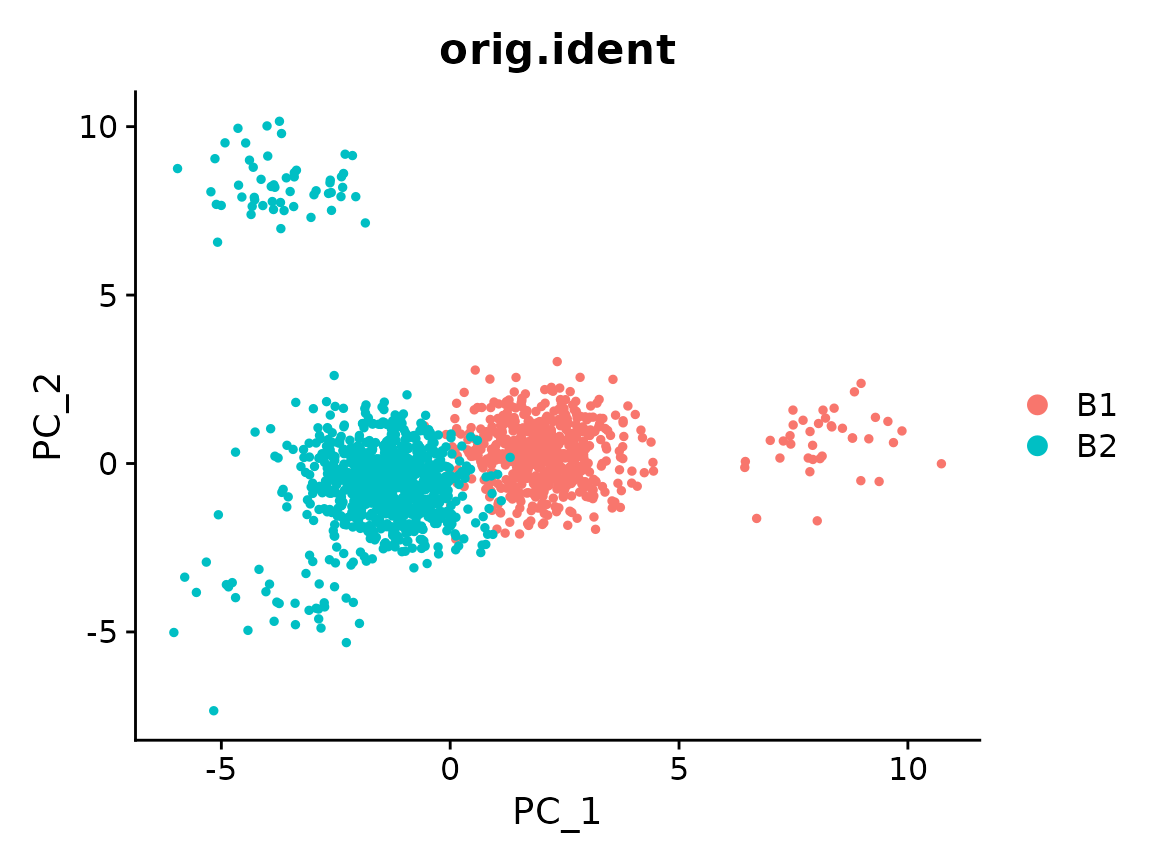
table(x$orig.ident)
#>
#> B1 B2
#> 631 948
x <- NormalizeData(x)
x <- FindVariableFeatures(x, nfeatures=100)
VariableFeaturePlot(x)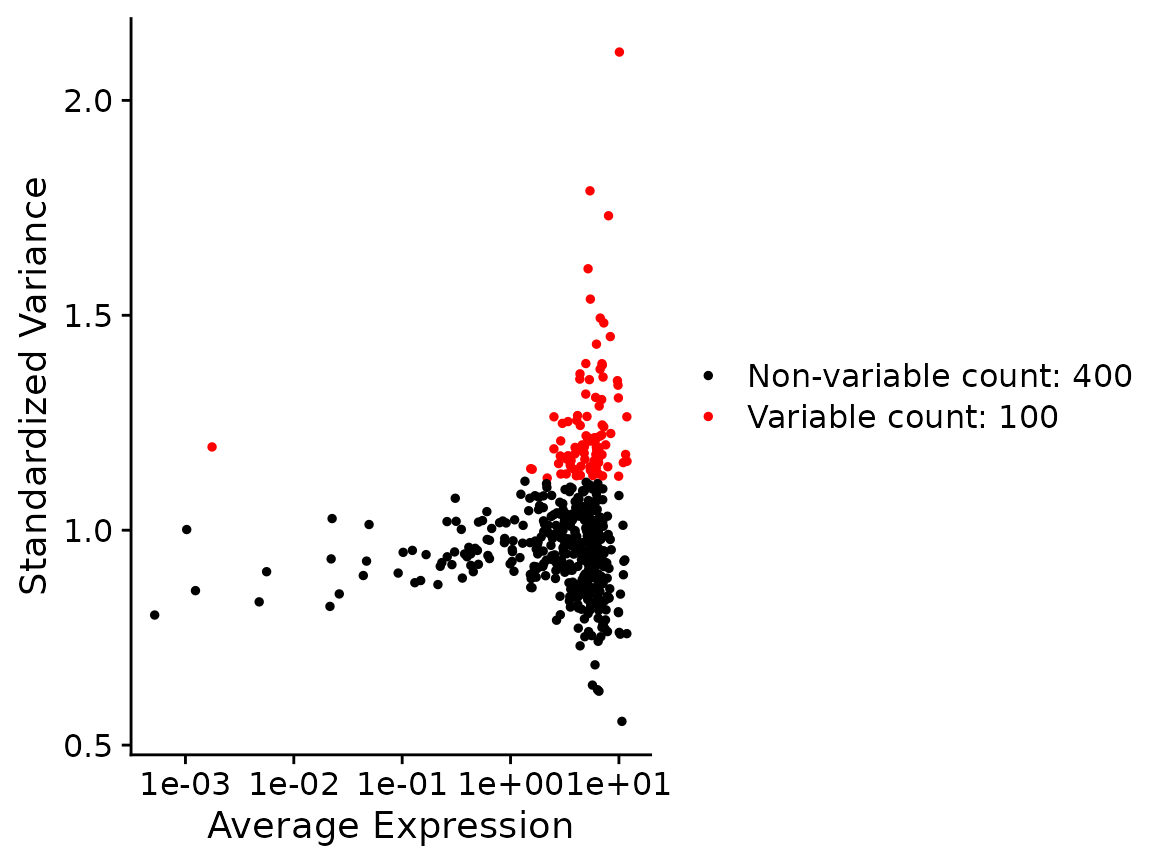
x <- ScaleData(x)
#> Centering and scaling data matrix
x <- RunPCA(x)
#> Warning in irlba(A = t(x = object), nv = npcs, ...): You're computing too large
#> a percentage of total singular values, use a standard svd instead.
#> PC_ 1
#> Positive: Gene130, Gene410, Gene373, Gene65, Gene274, Gene481, Gene285, Gene449, Gene382, Gene281
#> Gene323, Gene376, Gene229, Gene489, Gene473, Gene61, Gene50, Gene427, Gene174, Gene484
#> Gene193, Gene207, Gene24, Gene342, Gene297, Gene471, Gene292, Gene255, Gene226, Gene348
#> Negative: Gene461, Gene341, Gene492, Gene389, Gene178, Gene136, Gene468, Gene183, Gene366, Gene362
#> Gene234, Gene127, Gene416, Gene290, Gene5, Gene228, Gene311, Gene31, Gene9, Gene162
#> Gene370, Gene172, Gene474, Gene74, Gene386, Gene30, Gene401, Gene296, Gene326, Gene185
#> PC_ 2
#> Positive: Gene322, Gene31, Gene498, Gene297, Gene174, Gene223, Gene472, Gene228, Gene461, Gene341
#> Gene481, Gene489, Gene9, Gene449, Gene61, Gene176, Gene234, Gene160, Gene220, Gene427
#> Gene323, Gene373, Gene57, Gene303, Gene24, Gene410, Gene292, Gene298, Gene239, Gene382
#> Negative: Gene358, Gene17, Gene4, Gene229, Gene484, Gene296, Gene74, Gene366, Gene386, Gene65
#> Gene165, Gene213, Gene473, Gene226, Gene94, Gene369, Gene401, Gene185, Gene136, Gene376
#> Gene389, Gene78, Gene183, Gene492, Gene28, Gene36, Gene311, Gene285, Gene471, Gene326
#> PC_ 3
#> Positive: Gene178, Gene290, Gene255, Gene303, Gene449, Gene489, Gene481, Gene160, Gene239, Gene285
#> Gene61, Gene94, Gene366, Gene162, Gene136, Gene213, Gene342, Gene416, Gene369, Gene5
#> Gene24, Gene4, Gene17, Gene74, Gene226, Gene461, Gene473, Gene229, Gene390, Gene474
#> Negative: Gene183, Gene484, Gene472, Gene471, Gene322, Gene401, Gene296, Gene193, Gene386, Gene327
#> Gene185, Gene382, Gene223, Gene427, Gene3, Gene373, Gene174, Gene281, Gene274, Gene477
#> Gene305, Gene341, Gene31, Gene130, Gene311, Gene220, Gene207, Gene389, Gene498, Gene297
#> PC_ 4
#> Positive: Gene427, Gene477, Gene57, Gene322, Gene311, Gene183, Gene28, Gene22, Gene127, Gene30
#> Gene165, Gene5, Gene432, Gene474, Gene3, Gene239, Gene24, Gene281, Gene352, Gene390
#> Gene193, Gene65, Gene369, Gene78, Gene213, Gene50, Gene342, Gene354, Gene298, Gene292
#> Negative: Gene386, Gene274, Gene228, Gene94, Gene395, Gene327, Gene178, Gene223, Gene341, Gene449
#> Gene145, Gene461, Gene484, Gene468, Gene61, Gene473, Gene31, Gene297, Gene36, Gene498
#> Gene481, Gene234, Gene172, Gene185, Gene410, Gene290, Gene18, Gene207, Gene472, Gene174
#> PC_ 5
#> Positive: Gene327, Gene191, Gene36, Gene223, Gene162, Gene305, Gene220, Gene474, Gene50, Gene285
#> Gene74, Gene342, Gene174, Gene160, Gene234, Gene326, Gene373, Gene341, Gene468, Gene78
#> Gene382, Gene473, Gene352, Gene281, Gene416, Gene255, Gene375, Gene298, Gene94, Gene30
#> Negative: Gene178, Gene311, Gene297, Gene303, Gene410, Gene165, Gene461, Gene193, Gene471, Gene323
#> Gene239, Gene376, Gene65, Gene492, Gene386, Gene207, Gene31, Gene136, Gene472, Gene185
#> Gene481, Gene322, Gene362, Gene389, Gene489, Gene366, Gene484, Gene229, Gene427, Gene9Run Canek
We pass the column containing the batch information.
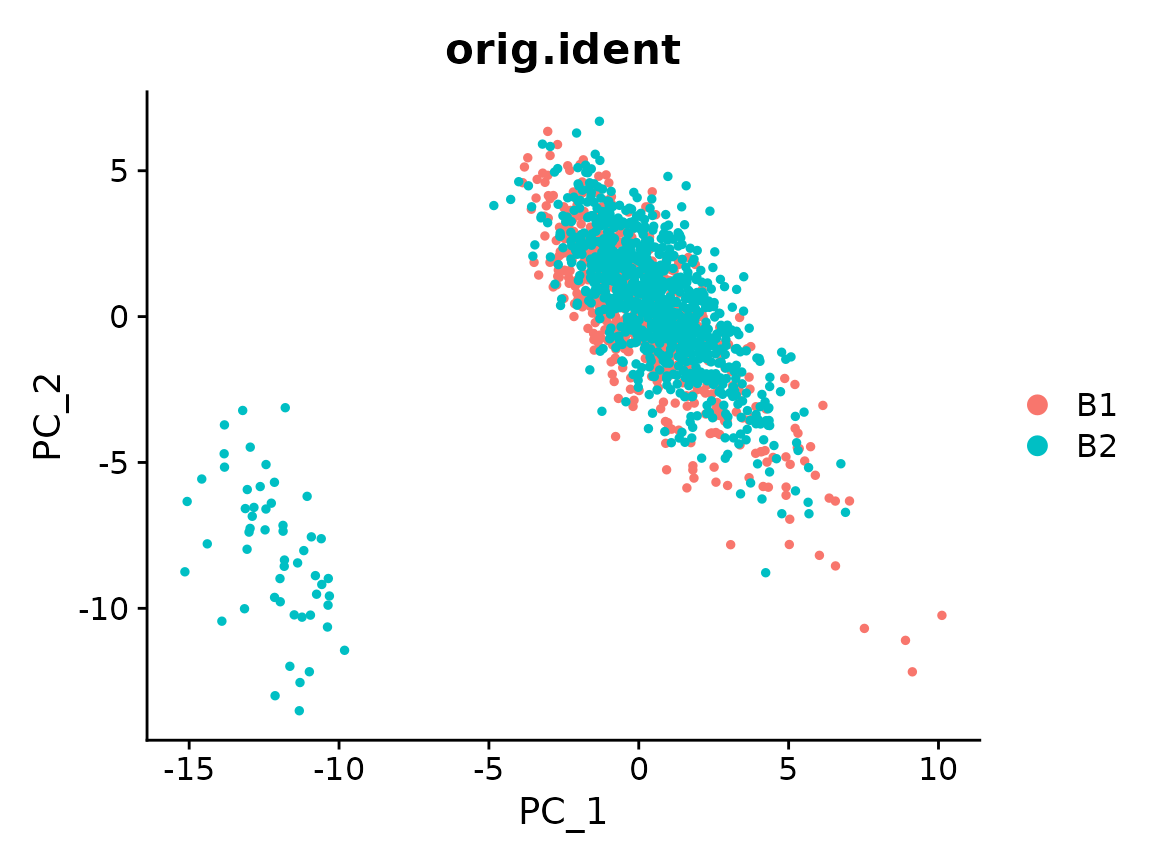
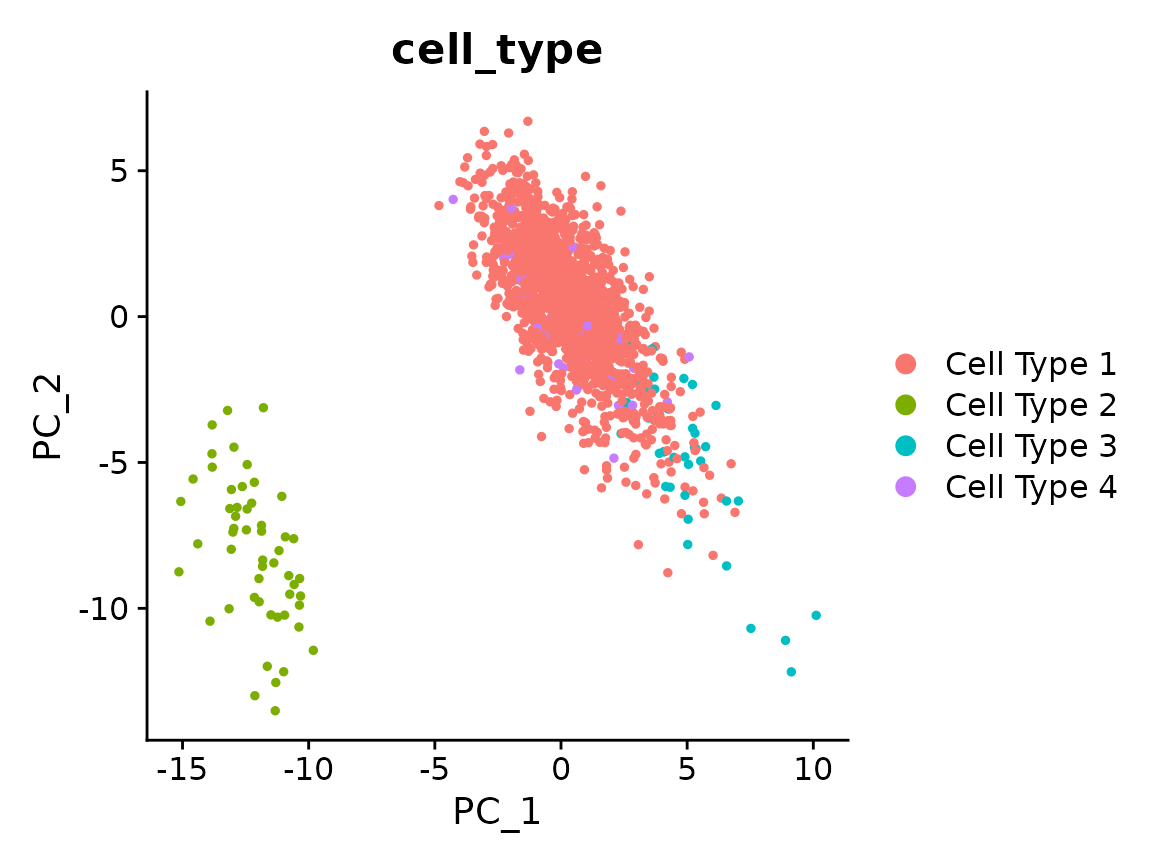
x <- RunCanek(x, "orig.ident")The features selected during integration are assigned as variable features.
head(VariableFeatures(x, assay="Canek"))
#> [1] "Gene348" "Gene322" "Gene57" "Gene461" "Gene61" "Gene145"We scale features and run PCA.
x <- ScaleData(x)
#> Centering and scaling data matrix
x <- RunPCA(x)
#> PC_ 1
#> Positive: Gene358, Gene150, Gene229, Gene488, Gene17, Gene484, Gene65, Gene169, Gene4, Gene392
#> Gene60, Gene387, Gene374, Gene299, Gene274, Gene180, Gene386, Gene167, Gene388, Gene96
#> Gene473, Gene454, Gene74, Gene338, Gene417, Gene285, Gene394, Gene94, Gene476, Gene205
#> Negative: Gene322, Gene461, Gene31, Gene202, Gene211, Gene472, Gene9, Gene341, Gene210, Gene118
#> Gene498, Gene297, Gene280, Gene122, Gene12, Gene325, Gene58, Gene2, Gene198, Gene111
#> Gene377, Gene64, Gene393, Gene168, Gene173, Gene480, Gene197, Gene456, Gene223, Gene300
#> PC_ 2
#> Positive: Gene164, Gene265, Gene366, Gene273, Gene17, Gene14, Gene4, Gene422, Gene237, Gene277
#> Gene213, Gene78, Gene126, Gene165, Gene399, Gene299, Gene374, Gene488, Gene156, Gene21
#> Gene85, Gene438, Gene286, Gene5, Gene147, Gene10, Gene319, Gene315, Gene40, Gene250
#> Negative: Gene274, Gene391, Gene476, Gene60, Gene76, Gene333, Gene295, Gene491, Gene339, Gene157
#> Gene88, Gene386, Gene490, Gene341, Gene31, Gene2, Gene228, Gene217, Gene223, Gene410
#> Gene322, Gene69, Gene459, Gene297, Gene472, Gene287, Gene449, Gene327, Gene133, Gene461
#> PC_ 3
#> Positive: Gene178, Gene102, Gene290, Gene490, Gene356, Gene449, Gene481, Gene303, Gene461, Gene394
#> Gene489, Gene262, Gene133, Gene67, Gene255, Gene345, Gene459, Gene307, Gene180, Gene446
#> Gene377, Gene142, Gene397, Gene189, Gene113, Gene239, Gene33, Gene73, Gene61, Gene118
#> Negative: Gene183, Gene391, Gene484, Gene179, Gene8, Gene1, Gene224, Gene401, Gene296, Gene471
#> Gene112, Gene312, Gene411, Gene476, Gene373, Gene327, Gene361, Gene382, Gene427, Gene107
#> Gene400, Gene437, Gene188, Gene472, Gene322, Gene193, Gene15, Gene87, Gene217, Gene132
#> PC_ 4
#> Positive: Gene461, Gene459, Gene492, Gene311, Gene178, Gene113, Gene386, Gene118, Gene389, Gene165
#> Gene132, Gene73, Gene133, Gene377, Gene183, Gene307, Gene198, Gene245, Gene164, Gene136
#> Gene335, Gene400, Gene366, Gene185, Gene411, Gene296, Gene484, Gene401, Gene471, Gene177
#> Negative: Gene449, Gene285, Gene60, Gene107, Gene373, Gene327, Gene189, Gene102, Gene402, Gene360
#> Gene274, Gene101, Gene345, Gene481, Gene489, Gene89, Gene174, Gene142, Gene160, Gene224
#> Gene281, Gene387, Gene342, Gene61, Gene67, Gene446, Gene223, Gene50, Gene49, Gene382
#> PC_ 5
#> Positive: Gene427, Gene112, Gene33, Gene322, Gene281, Gene198, Gene477, Gene15, Gene358, Gene8
#> Gene108, Gene485, Gene183, Gene57, Gene394, Gene449, Gene490, Gene111, Gene481, Gene102
#> Gene180, Gene250, Gene290, Gene489, Gene406, Gene434, Gene411, Gene178, Gene455, Gene140
#> Negative: Gene386, Gene306, Gene228, Gene94, Gene190, Gene20, Gene157, Gene224, Gene60, Gene473
#> Gene333, Gene356, Gene383, Gene274, Gene338, Gene302, Gene395, Gene156, Gene175, Gene223
#> Gene270, Gene137, Gene321, Gene19, Gene456, Gene45, Gene78, Gene79, Gene181, Gene472